Nucleome Therapeutics: Decoding the Regulatory Genome
In the vast landscape of human genetics, only 2-3% of our DNA directly codes for proteins—the building blocks of life. The remaining 97-98% was once dismissed as “junk DNA” but is now understood to be crucial regulatory code that controls how, when, and where genes are expressed.
Oxford University spin-out Nucleome Therapeutics is pioneering a revolutionary approach to decode this regulatory genome, with profound implications for how we understand and treat human diseases. Their platform, which combines sophisticated machine learning with next-generation sequencing technology, is already yielding promising drug candidates for autoimmune conditions like rheumatoid arthritis and lupus.
Understanding the True Causes of Disease
“The fundamental problem in developing medicines is understanding the complex biology of disease and what goes wrong when people get diseases,” explains Dr. Mark Bodmer, CEO of Nucleome Therapeutics.
Traditional drug discovery faces a significant translation problem. Scientists have become “great mouse doctors,” as Bodmer puts it, developing treatments that work well in animal models but often fail when tested in humans. The disconnect stems from focusing primarily on protein-coding genes while overlooking the critical regulatory elements that differ dramatically between species.
The human genome contains approximately three billion base pairs, with genetic variations scattered throughout. Some of these variations are associated with increased disease risk, but identifying which ones actually cause disease—and how—has been extraordinarily difficult. This is especially true because most disease-associated variants don’t alter proteins directly; instead, they modify how genes are regulated.
“90% or more of those mutations occur in what’s called non-coding genetics,” explains Dr. Stephen Harrison, CSO at Nucleome Therapeutics. “How on earth do you figure out what that mutation means in terms of a disease mechanism?”
This challenge is illustrated by diseases like rheumatoid arthritis, where approximately 49,000 genetic variants have been associated with the disease, but determining which variants actually cause disease has been nearly impossible—until now.
Reading the Genome in 3D
Nucleome’s breakthrough lies in its ability to analyze the three-dimensional structure of DNA rather than just its linear sequence. The regulatory genome can’t be understood in one dimension because enhancers (genetic switches) can be positioned hundreds of thousands or even millions of base pairs away from the genes they control in the linear DNA sequence. These distant elements interact through the folding of DNA in three-dimensional space within the cell nucleus.
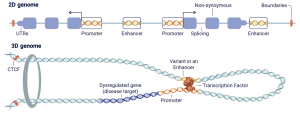
From 2‑D to 3‑D: how distant regulatory DNA contacts its target gene and why the 3‑D view matters. In the linear (2‑D) genome, promoters, exons, and distal enhancers sit far apart along the DNA strand. Inside the nucleus, however, the same stretch of DNA is folded into chromatin loops anchored by CTCF (an architectural protein). This looping brings a far‑away enhancer into physical contact with its promoter, allowing bound transcription factors to switch the gene on. A single‑nucleotide variant inside the enhancer (shown in orange) can weaken or strengthen the transcription‑factor binding site, mis‑regulate the target gene (dark‑blue exons) and drive disease. Nucleome’s technology maps these long‑range contacts at single‑base resolution, enabling researchers to link non‑coding variants to the genes they actually control.
Dr. Jonny Wray, CTO at Nucleome Therapeutics, explains their core technologies: “We focus on the machine learning first. What we do there is ask the question of all of the human genetic variation that we can measure and associate with disease, which ones are the ones that are actually doing something, because most of what you end up measuring is effectively statistical noise.”
This machine learning approach narrows down hundreds of thousands of genetic variants to a more manageable number that can be experimentally tested. Their proprietary functional genomics technology (called MCC) then determines how regions that are far apart in the linear genome interact when DNA folds in three dimensions, with single base-pair resolution—the only technology able to achieve this precision.
“We link disease to variants, to genes in specific cell types,” says Wray. “And when we can do that genome-wide, we can start to ask questions: What is the underlying mechanistic dysregulation? What is going wrong in autoimmune disease, and therefore how can we treat them?”
The scale of this approach is impressive—Nucleome has analyzed more than ~155k SNPs (single nucleotide polymorphisms) across 17 different cell types, including rare immune cell types and activation models crucial for understanding autoimmune diseases. Their technology has examined hundreds of thousands of interactions, with recent improvements enabling a dramatic increase in throughput. According to Nucleome’s research data, this approach increases the probability of clinical success by ~3.5 times compared to traditional (non-genetic) drug discovery methods.
Targeting Inflammation Resolution: A New Frontier
Nucleome is applying its platform to inflammatory and autoimmune diseases, including rheumatoid arthritis, lupus, multiple sclerosis, and inflammatory bowel disease. Their approach has identified entirely new targets and mechanisms.
“All of the drugs that we currently have actually block what are called the pro-inflammatory mediators on the up phase,” Bodmer explains. “So some of them are quite effective, but they’re not actually switching the thing off. They’re just sort of catching the water as it comes out of the tap rather than turning the tap off.”
Nucleome’s lead program targets what they call an “inflammation checkpoint”—a protein that naturally helps resolve inflammation. Using their genetic analysis, they discovered that lower levels of this protein are associated with higher risk of autoimmune diseases, suggesting its important role in turning inflammation off.
“There’s a half the immune system, a whole half of the molecular biology of the system, which is untapped from a drug point of view,” notes Bodmer. By developing antibodies that stimulate this checkpoint, Nucleome aims to promote the natural resolution of inflammation rather than simply suppressing it.
From Discovery to Development: NTP464 for Rheumatoid Arthritis
Nucleome’s approach has already yielded promising drug targets, with their lead candidate NTP464 advancing through preclinical development for rheumatoid arthritis.
This target was identified when their machine learning algorithm predicted a strong loss-of-function variant associated with rheumatoid arthritis risk. Through their 3D genome technology, they discovered this variant destroys an enhancer-promoter link specifically in B cells, decreasing expression of this critical protein.
What makes NTP464 particularly interesting is that it represents a novel approach to treating autoimmune disease. Rather than suppressing inflammation directly, an agonist antibody targeting NTP464 promotes natural anti-inflammatory activities. In preclinical studies, this approach reduced disease severity in a collagen-induced arthritis model, demonstrating its potential clinical value.
Beyond Single Targets: Understanding Disease Systems
The scale of Nucleome’s technology allows them to go beyond identifying individual drug targets to understanding entire disease mechanisms. This enables patient stratification—identifying subgroups within a disease that may respond differently to treatments.
“Rheumatoid arthritis isn’t actually one disease when you think of it from a mechanistic point of view,” says Dr. Wray. “It could be multiple mechanisms that drive the disease. And the consequences of that is an individual with RA may or may not respond to a drug, because you’re actually drugging a mechanism, not a disease, not a symptom.”
By clustering genetic variants around specific immune mechanisms, Nucleome can define “endotypes” that represent patient subgroups with different molecular pathologies. This has practical implications for drug development—their analysis of NTP464 revealed connections to specific biological pathways, including tryptophan/kynurenine metabolism, that would have been difficult to discover through traditional means.
The Future: From Understanding to Treatment
Nucleome is at a milestone point with their lead candidate antibody entering preclinical development. They plan to conduct early human studies to validate whether the mechanisms observed in healthy cells also work in patient cells.
Their pipeline shows impressive progress: from analyzing 428 genes mapped to disease, they’ve prioritized 281 targets, with 11 targets having expression validation, 5 with biology validation, and NTP464 advancing into antibody development. Additionally, they’ve identified 36 non-antibody targets that could be addressed with alternative therapeutic approaches.
Beyond their current pipeline, the company continues to refine its genetic analysis technology and build an “exponentially growing database of gene control in health and disease.” Their long-term vision includes identifying novel targets within the regulatory genome itself that could be modified through genetic medicines.
“Understanding human disease is in humans,” emphasizes Bodmer, paraphrasing Alexander Pope’s famous line. By focusing on human genetics from the start, Nucleome aims to fundamentally transform how we develop new treatments—turning the “architecture plans” of our DNA into blueprints for healing.